GenAI diffusion models learn to generate new content more consistently than expected
Artificial intelligence (AI) image generators have taken the world by storm in recent years, as users plaster social media with artificially-generated artwork, and popular software companies such as Adobe build generative AI tools into their programs.
A team at the University of Michigan has uncovered new insights into the ability of a generative AI technique known as a diffusion model to produce nearly identical contents even under different training settings. The research could be applied to a better understanding of GenAI techniques, while also being used to safeguard users against adversarial attacks.
“Our result provides new light into the working mechanism of diffusion models,” said Qing Qu, assistant professor in Electrical and Computer Engineering, “We demonstrated that the generative capability of diffusion models stems from their ability to learn the true underlying distribution.”

Diffusion models are trained through a process that begins with sharp, clear images, to which noise is incrementally added, progressively blurring the image and introducing variation. This noise manifests in the images as a grainy pattern, resembling the static seen on an old television set without reception. The core learning mechanism of diffusion models involves reversing this process, systematically removing the noise in a step-by-step “denoising” process to recreate a clear image.
The capacity of various diffusion models to create similar content from the same noise input is recognized as reproducibility. The team from the University of Michigan has, for the first time, provided a comprehensive study of this phenomenon, showing compelling evidence that diffusion models are superior in learning the underlying data distribution compared to earlier models.
More interestingly, the team found out that the model reproducibility manifests in two distinct training regimes. First, there’s a “memorization regime,” where the model attempts to remember everything it sees in the data it’s trained on. Second, there’s a “generalization regime,” where the model starts to learn the overall patterns in the data, instead of merely memorizing it.
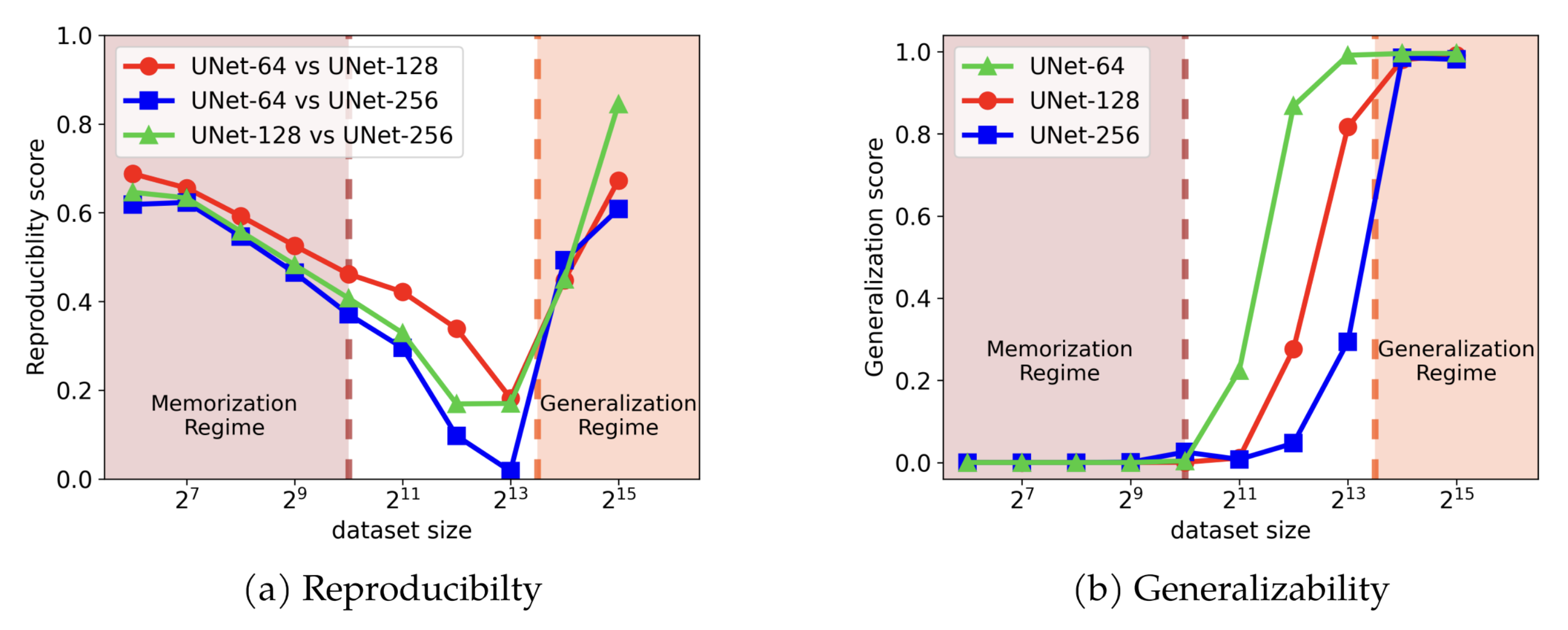
“We observe reproducibility in diffusion models within both of these regimes, yet they converge towards two distinct data distributions.” said Huijie Zhang, PhD student in ECE.
While the reproducibility in the memorization regime seems intuitive, the reproducibility in the generalization regime has not been well-studied. In the worst case, explained Zhang, >213 training samples were needed to interpolate and extrapolate the image distribution, but the results showed that the actual samples are much smaller, which was quite striking.
Researchers will now focus on understanding how the diffusion models can generate content that is both novel and consistently similar across different models.
“Our work provides convincing evidence that diffusion models are able to learn the underlying distribution of the data very well under various settings in a robust manner.” said Prof. Liyue Shen, who is collaborating on the project. “This will open up many exciting research directions for generative AI in the near future including for medical and scientific applications in general.”
For example, by leveraging the reproducibility across different noise levels in diffusion models, it may allow researchers to increase the accuracy and efficiency of other types of generative models, including audio, video, and scientific applications.
Moreover, the findings may also be applied to decrease instances of adversarial attacks, in which a model is provided with deceptive data that prompts it to make an incorrect (e.g., providing an image of an orange cat for the prompt “goldfish”) or inappropriate (e.g., providing information on how to commit a crime) response.
The research, The Emergence of Reproducibility and Consistency in Diffusion Models, was presented at the NeurIPS 2023 Workshop on Diffusion Models, where it received the Best Paper Award. The paper was co-first authored by Huijie Zhang and Jinfan Zhou, a master’s student in Robotics. Yifu Lu and Minzhe Guo, undergraduate students in Computer Science and Engineering, contributed to the experimental work for this project.
The project was supported by funding from the National Science Foundation (NSF), the Office of Naval Research, Amazon Web Services, the Michigan Institute for Computational Discovery & Engineering and the Michigan Institute for Data Science.